Global Terrorist Attacks
Here Global Terrorism Database (GTD) is used to classify the Terrorist Group which might be involved in an attack based on number of kills, region, weapons used etc.
The tutorial contains a jupyter notebook, which is used to preprocess the data and classify the Terrorist Groups using Machine Learning techniques like Support Vector Machine and Decision Tree with accuracies of 96.55% and 98.27% respectively.
The input and labels of the model are stored in the x_normed.npy and y.npy files respectively.
Further details about the Database can be found in Codebook.pdf file.
import pandas as pd
import numpy as np
import pickle, gzip
import matplotlib.pyplot as plt
from sklearn import svm
from sklearn.metrics import confusion_matrix, precision_recall_fscore_support
from sklearn.tree import DecisionTreeClassifier
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
Removing NA values
df = pd.read_excel('globalterrorismdb_0718dist.xlsx') # reading all data
df.shape
(181691, 135)
df.head()
| iyear | imonth | iday | approxdate | extended | resolution | country | ... | dbsource | INT_LOG | INT_IDEO | INT_MISC | INT_ANY | related | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1970 | 7 | 2 | NaN | 0 | NaT | 58 | ... | PGIS | 0 | 0 | 0 | 0 | NaN |
| 1 | 1970 | 0 | 0 | NaN | 0 | NaT | 130 | ... | PGIS | 0 | 1 | 1 | 1 | NaN |
| 2 | 1970 | 1 | 0 | NaN | 0 | NaT | 160 | ... | PGIS | -9 | -9 | 1 | 1 | NaN |
| 3 | 1970 | 1 | 0 | NaN | 0 | NaT | 78 | ... | PGIS | -9 | -9 | 1 | 1 | NaN |
| 4 | 1970 | 1 | 0 | NaN | 0 | NaT | 101 | ... | PGIS | -9 | -9 | 1 | 1 | NaN |
5 rows × 135 columns
df.isnull().any() # checking columns with null value
eventid False
iyear False
imonth False
iday False
approxdate True
extended False
resolution True
country False
...
propextent True
propextent_txt True
propvalue True
propcomment True
ishostkid True
nhostkid True
nhostkidus True
nhours True
INT_MISC False
INT_ANY False
related True
Length: 135, dtype: bool
# To remove all columns having null values > 5000
for i in df.columns:
if df[i].isnull().sum()>5000:
df.drop(columns=i, inplace=True)
df.shape
(181691, 41)
# further reduction of unnecessary columns after looking at the Codebook (documentation)
df.drop(columns=['eventid', 'extended', 'country_txt', 'region_txt', 'specificity', 'vicinity',
'crit1', 'crit2', 'crit3', 'attacktype1_txt', 'targtype1_txt', 'natlty1_txt',
'guncertain1', 'individual', 'weaptype1_txt', 'dbsource', 'INT_MISC'], inplace=True)
df.shape
(181691, 24)
# saving file for future use
df.to_csv('terrorist_data_pruned.csv', index=False)
Filling NA values
df2 = pd.read_csv('terrorist_data_pruned.csv')
df2.head()
| iyear | imonth | iday | country | region | provstate | city | ... | natlty1 | gname | weaptype1 | property | ishostkid | INT_LOG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1970 | 7 | 2 | 58 | 2 | -1 | Santo Domingo | ... | 58.0 | MANO-D | 13 | 0 | 0.0 | 0 |
| 1 | 1970 | 0 | 0 | 130 | 1 | 0 | Mexico city | ... | 21.0 | 23rd of September Communist League | 13 | 0 | 1.0 | 0 |
| 2 | 1970 | 1 | 0 | 160 | 5 | 1 | Unknown | ... | 217.0 | Unknown | 13 | 0 | 0.0 | -9 |
| 3 | 1970 | 1 | 0 | 78 | 8 | 2 | Athens | ... | 217.0 | Unknown | 6 | 1 | 0.0 | -9 |
| 4 | 1970 | 1 | 0 | 101 | 4 | 3 | Fukouka | ... | 217.0 | Unknown | 8 | 1 | 0.0 | -9 |
5 rows × 24 columns
# replacing nan values (refer Codebook.pdf)
df2['doubtterr'].fillna(-9, inplace=True)
df2['multiple'].fillna(0, inplace=True)
df2['natlty1'].fillna(0, inplace=True)
df2['ishostkid'].fillna(-9, inplace=True)
df2['latitude'].fillna(0, inplace=True)
df2['longitude'].fillna(0, inplace=True)
# quantizing dataset
df2['provstate'] = pd.factorize(df2['provstate'])[0]
df2['iyear'] = pd.factorize(df2['iyear'])[0]
df2['city'] = pd.factorize(df2['city'])[0]
df2['country'] = pd.factorize(df2['country'])[0]
df2['target1'] = pd.factorize(df2['target1'])[0]
df2['gname'] = pd.factorize(df2['gname'])[0]
df2['natlty1'] = pd.factorize(df2['natlty1'])[0]
df2.isna().any()
iyear False
imonth False
iday False
country False
region False
provstate False
city False
latitude False
longitude False
doubtterr False
multiple False
success False
suicide False
attacktype1 False
targtype1 False
target1 False
natlty1 False
gname False
weaptype1 False
property False
ishostkid False
INT_LOG False
INT_IDEO False
INT_ANY False
dtype: bool
# saving file for future use
df2.to_csv('terrorist_data_pruned.csv', index=False)
Normalizing Columns
df = pd.read_csv('terrorist_data_pruned.csv')
df.head()
| iyear | imonth | iday | country | region | provstate | city | latitude | ... | natlty1 | gname | weaptype1 | property | ishostkid | INT_LOG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 7 | 2 | 0 | 2 | 0 | 0 | 18.456792 | ... | 0 | 0 | 13 | 0 | 0.0 | 0 |
| 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 19.371887 | ... | 1 | 1 | 13 | 0 | 1.0 | 0 |
| 2 | 0 | 1 | 0 | 2 | 5 | 2 | 2 | 15.478598 | ... | 2 | 2 | 13 | 0 | 0.0 | -9 |
| 3 | 0 | 1 | 0 | 3 | 8 | 3 | 3 | 37.997490 | ... | 2 | 2 | 6 | 1 | 0.0 | -9 |
| 4 | 0 | 1 | 0 | 4 | 4 | 4 | 4 | 33.580412 | ... | 2 | 2 | 8 | 1 | 0.0 | -9 |
5 rows × 24 columns
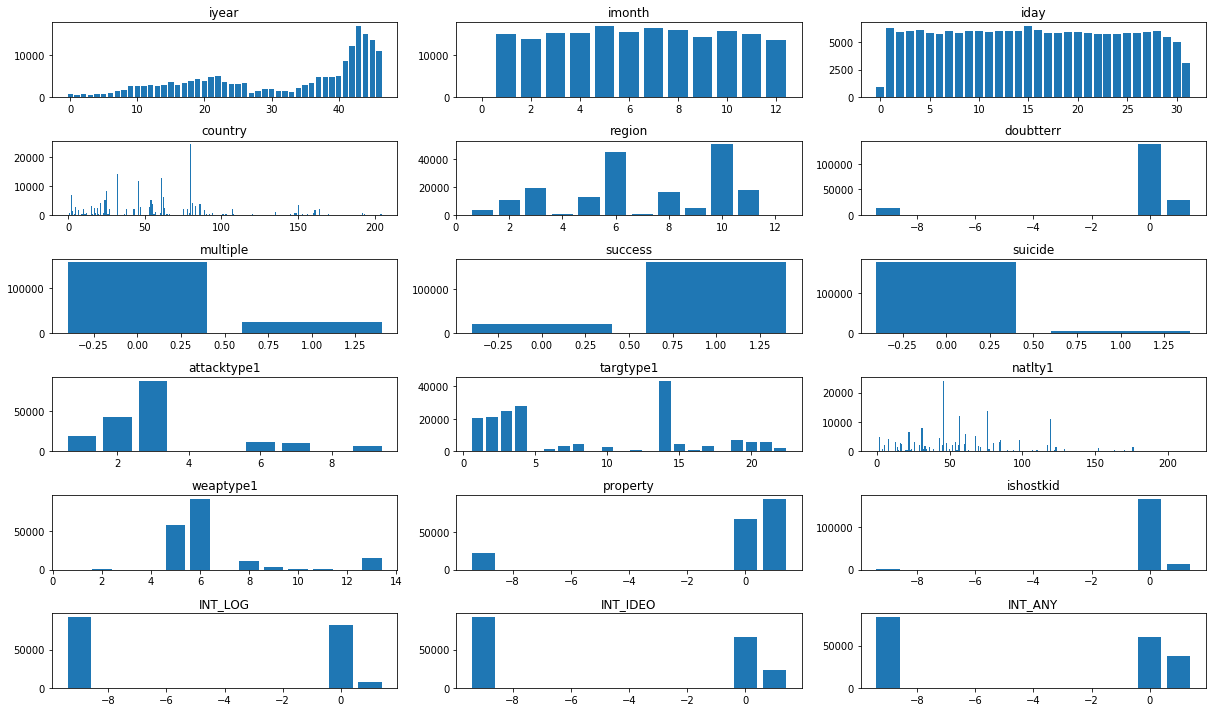
# checking frequency of column values
plt.figure(figsize=(17,10))
i = 0
for col in df.columns:
if col=='latitude' or col=='longitude' or col=='city' or col=='provstate' or col=='target1' or col=='gname':
continue
i += 1
plt.subplot(6, 3, i)
num_list = df[col].value_counts()
plt.bar(num_list.index, num_list.values)
plt.title(col)
plt.tight_layout()
plt.show()
# classes having less than 600 samples were dropped from the dataframe to remove skewedness in data
num = df['gname'].value_counts()
res = num.where(num>600).dropna()
res
2 82782.0
2001 7478.0
3024 5613.0
455 4555.0
478 3351.0
2699 3288.0
15 2772.0
46 2671.0
203 2487.0
2819 2418.0
1057 2310.0
78 2024.0
2589 1878.0
631 1630.0
221 1606.0
123 1561.0
2695 1351.0
36 1125.0
2632 1062.0
2549 1020.0
948 895.0
1019 830.0
863 716.0
157 639.0
2526 638.0
598 632.0
3136 624.0
281 607.0
Name: gname, dtype: float64
k = []
c = 0
for index, row in df.iterrows():
if row['gname'] not in list(res.index):
k.append(index)
if row['gname']==2:
c += 1
if c>5000:
k.append(index)
df.drop(index=k, inplace=True)
df.shape
(60781, 24)
# converts labels to one-hot encoding
y = pd.get_dummies(df['gname']).values
y.shape
(60781, 28)
# normalizing feature values
x = df.drop(columns='gname')
x = x.values
min_max_scaler = MinMaxScaler()
x_scaled = min_max_scaler.fit_transform(x)
df2 = pd.DataFrame(x_scaled)
x_scaled.shape
(60781, 23)
df2.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ... | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.083333 | 0.000000 | 0.010 | 0.363636 | 0.000708 | 0.000082 | 0.572701 | ... | 1.000000 | 0.9 | 0.9 | 0.0 | 0.0 | 1.0 |
| 1 | 0.0 | 0.083333 | 0.000000 | 0.015 | 0.636364 | 0.001062 | 0.000109 | 0.782926 | ... | 0.363636 | 1.0 | 0.9 | 0.0 | 0.0 | 1.0 |
| 2 | 0.0 | 0.083333 | 0.000000 | 0.020 | 0.272727 | 0.001416 | 0.000136 | 0.741690 | ... | 0.545455 | 1.0 | 0.9 | 0.0 | 0.0 | 1.0 |
| 3 | 0.0 | 0.083333 | 0.064516 | 0.025 | 0.000000 | 0.002478 | 0.000218 | 0.781007 | ... | 0.363636 | 1.0 | 0.9 | 0.0 | 0.0 | 0.0 |
| 4 | 0.0 | 0.083333 | 0.258065 | 0.035 | 0.636364 | 0.003540 | 0.000327 | 0.819274 | ... | 0.272727 | 0.9 | 0.9 | 0.0 | 0.0 | 1.0 |
5 rows × 23 columns
# saving file for future use
np.save('x_normed.npy', x_scaled)
np.save('y.npy', y)
Classification
x = np.load('x_normed.npy')
y = np.load('y.npy')
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.1, random_state=0)
print(x_train.shape, y_train.shape, x_test.shape, y_test.shape)
(54702, 23) (54702, 28) (6079, 23) (6079, 28)
model1 = svm.SVC(C=5, kernel='rbf')
model2 = DecisionTreeClassifier()
model1.fit(x_train, np.argmax(y_train, axis=1))
SVC(C=5, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='auto', kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)
model2.fit(x_train, np.argmax(y_train, axis=1))
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False, random_state=None,
splitter='best')
pred1 = model1.predict(x_test)
acc1 = np.mean(pred1==np.argmax(y_test, axis=1))*100
pred2 = model2.predict(x_test)
acc2 = np.mean(pred2==np.argmax(y_test, axis=1))*100
print('Accuracy SVC: {:.2f}%\nAccuracy Desicion Tree: {:.2f}%'.format(acc1, acc2))
Accuracy SVC: 96.55%
Accuracy Desicion Tree: 98.27%
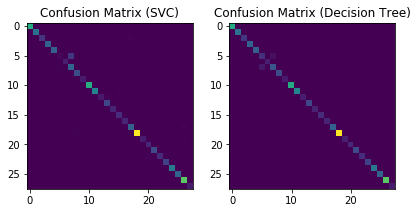
cm1 = confusion_matrix(np.argmax(y_test, axis=1), pred1)
cm2 = confusion_matrix(np.argmax(y_test, axis=1), pred2)
plt.subplot(1, 2, 1)
plt.imshow(cm1)
plt.title('Confusion Matrix (SVC)')
plt.subplot(1, 2, 2)
plt.imshow(cm2)
plt.title('Confusion Matrix (Decision Tree)')
plt.tight_layout()
plt.show()
pr1 = precision_recall_fscore_support(np.argmax(y_test, axis=1), pred1)
pr2 = precision_recall_fscore_support(np.argmax(y_test, axis=1), pred2)
print('SVC\nPrecision: {:.2f}%\nRecall: {:.2f}%\nF-score: {:.2f}%'.format(np.mean(pr1[0])*100, np.mean(pr1[1])*100, np.mean(pr1[2])*100))
print('\nDecision Tree\nPrecision: {:.2f}%\nRecall: {:.2f}%\nF-score: {:.2f}%'.format(np.mean(pr2[0])*100, np.mean(pr2[1])*100, np.mean(pr2[2])*100))
SVC
Precision: 96.12%
Recall: 94.68%
F-score: 94.52%
Decision Tree
Precision: 97.76%
Recall: 97.95%
F-score: 97.84%